Please join us for a free live webinar with guest speaker Dr. József Kardos
March 12, 2025 at 1:00 pm EDT
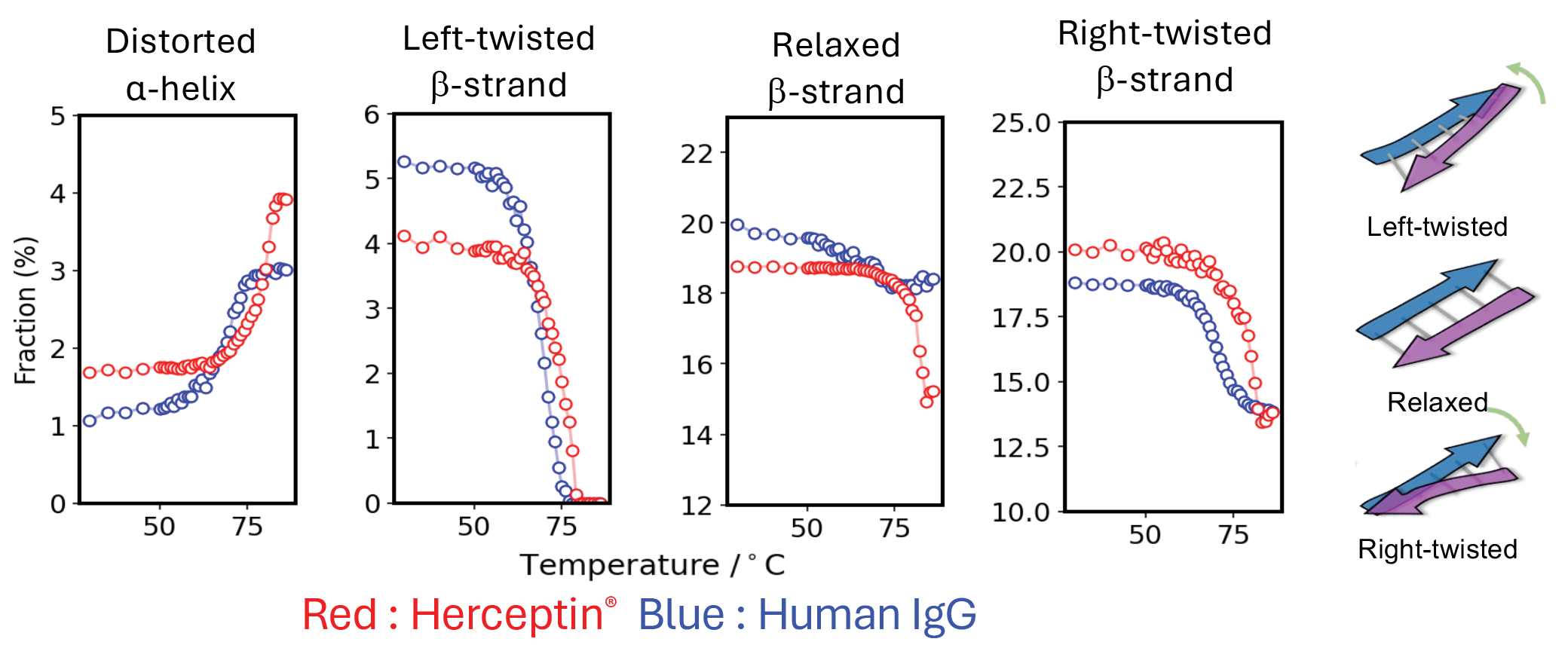 Proteins are fundamental to nearly all biological processes and serve as key targets in modern molecular biology, biotechnology, and the pharmaceutical industry. Understanding their three-dimensional structure is essential for deciphering their functions, manipulating them, or designing novel proteins to address economic, health, and environmental challenges. To bypass the traditional, time-intensive experimental approaches, AlphaFold was developed to accelerate the modelling of protein 3D structures. While AlphaFold typically delivers accurate predictions, it faces challenges in accounting for crucial factors such as pH, ionic strength, temperature, mutations, and post-translational modifications—parameters that are vital for drug design and ensuring product stability. Therefore, the integration of experimental techniques alongside computational modeling is essential for achieving complete reliability. Spectroscopic methods provide valuable insights into protein structure, fold, stability, and dynamics under various conditions. Circular dichroism (CD) spectroscopy, in particular, offers a powerful approach to characterizing protein secondary structures. When combined with the recently developed BeStSel method (https://bestsel.elte.hu), CD spectroscopy characterizes precisely protein secondary structures and predicts fold from a single measurement. BeStSel uniquely resolves the spectral diversity of β-structured proteins, a long-standing challenge in CD spectroscopy. Its user-friendly web server facilitates the analysis of both single and multiple CD spectra, offering an intuitive and intelligent interface. What sets BeStSel apart is its ability to distinguish eight secondary structure components, including parallel and antiparallel β-sheets with three distinct twist groups. It surpasses existing methods in both accuracy and information content and, uniquely predicts protein fold.
Proteins are fundamental to nearly all biological processes and serve as key targets in modern molecular biology, biotechnology, and the pharmaceutical industry. Understanding their three-dimensional structure is essential for deciphering their functions, manipulating them, or designing novel proteins to address economic, health, and environmental challenges. To bypass the traditional, time-intensive experimental approaches, AlphaFold was developed to accelerate the modelling of protein 3D structures. While AlphaFold typically delivers accurate predictions, it faces challenges in accounting for crucial factors such as pH, ionic strength, temperature, mutations, and post-translational modifications—parameters that are vital for drug design and ensuring product stability. Therefore, the integration of experimental techniques alongside computational modeling is essential for achieving complete reliability. Spectroscopic methods provide valuable insights into protein structure, fold, stability, and dynamics under various conditions. Circular dichroism (CD) spectroscopy, in particular, offers a powerful approach to characterizing protein secondary structures. When combined with the recently developed BeStSel method (https://bestsel.elte.hu), CD spectroscopy characterizes precisely protein secondary structures and predicts fold from a single measurement. BeStSel uniquely resolves the spectral diversity of β-structured proteins, a long-standing challenge in CD spectroscopy. Its user-friendly web server facilitates the analysis of both single and multiple CD spectra, offering an intuitive and intelligent interface. What sets BeStSel apart is its ability to distinguish eight secondary structure components, including parallel and antiparallel β-sheets with three distinct twist groups. It surpasses existing methods in both accuracy and information content and, uniquely predicts protein fold.
Here, we present a robust methodology for protein structure determination and highlight the crucial role of CD spectroscopy in validating bioinformatics predictions, using numerous examples of disease-related proteins. With desktop or cutting-edge plate reader CD devices, high-throughput experimental validation and protein structure screening become a reality. Moreover, BeStSel is now integrated into Spectra Manager 2.5 of JASCO’s new CD instruments, providing an advanced tool for industry and academia.
Click here to register for the live webinar March 12 at 1:00 pm.
 Dr. József Kardos obtained his MSc in Physics-Biophysics from Eötvös Loránd University in Budapest, Hungary, followed by a PhD in Structural Biochemistry from the Biology Doctoral School of the same university. From 2001 to 2004, he held a postdoctoral position at the Institute of Protein Research, Osaka University, Japan. In 2004, he returned to the Department of Biochemistry at Eötvös Loránd University. Since 2014, he has been leading the Neuroimmunology Research Group. His primary research interests include protein folding and misfolding, the structural and thermodynamic principles behind protein aggregation and amyloid formation, proteomics studies on the role of protein aggregation in neurodegeneration, immune complement system activation, and the complement system’s function in the brain. He has developed innovative methods to investigate the thermodynamics of amyloid formation and to analyze protein secondary structure and fold using CD spectroscopy. Together with his colleague Dr. András Micsonai, he developed the BeStSel method, which allows for the estimation of protein structure from CD spectra with superior accuracy. Their publications on the method have been cited in over two thousand scientific papers and has become integral to the curricula of numerous prestigious universities. Additionally, they created user-friendly software and established a web server for the global research community, which is currently used by thousands of scientists around the world.
Dr. József Kardos obtained his MSc in Physics-Biophysics from Eötvös Loránd University in Budapest, Hungary, followed by a PhD in Structural Biochemistry from the Biology Doctoral School of the same university. From 2001 to 2004, he held a postdoctoral position at the Institute of Protein Research, Osaka University, Japan. In 2004, he returned to the Department of Biochemistry at Eötvös Loránd University. Since 2014, he has been leading the Neuroimmunology Research Group. His primary research interests include protein folding and misfolding, the structural and thermodynamic principles behind protein aggregation and amyloid formation, proteomics studies on the role of protein aggregation in neurodegeneration, immune complement system activation, and the complement system’s function in the brain. He has developed innovative methods to investigate the thermodynamics of amyloid formation and to analyze protein secondary structure and fold using CD spectroscopy. Together with his colleague Dr. András Micsonai, he developed the BeStSel method, which allows for the estimation of protein structure from CD spectra with superior accuracy. Their publications on the method have been cited in over two thousand scientific papers and has become integral to the curricula of numerous prestigious universities. Additionally, they created user-friendly software and established a web server for the global research community, which is currently used by thousands of scientists around the world.
