Myoglobin Secondary Structure Measurement using Circular Dichroism with Automated pH Titration
August 24, 2022
Introduction
Effects of pH on Myoglobin Secondary Structure
In this application note the effects of pH on myoglobin secondary structure is measured using circular dichroism with automated titration. Circular dichroism spectroscopy (CD) is very sensitive to the secondary structure of poly-peptides and proteins. Circular dichroism is a form of light absorption spectroscopy that measures the difference in absorbance of right and left-circularly polarized light (rather than the commonly used absorbance of isotropic light) by a substance. It has been shown that CD spectra between 260 and approximately 180 nm can be analyzed for the different secondary structure types: alpha helix, beta sheet, beta turn, random coils, etc. Secondary structure determination by CD is reported to achieve accuracies of 0.97 for helices, 0.75 for beta sheet, 0.50 for turns, and 0.89 for other structural motifs.
A protein can be considered as a hetero-polymer, often hundreds of units long, composed of twenty basic amino acids. The sequence of amino acids dictates the well-defined native 3D structure, which often includes helix, sheet, turn, and random coil regions. Although the backbone of amino acids is covalently linked, usually non-covalent interactions are responsible for the folded 3D structure. In the native protein, the hydrophobic amino acid residues are generally sequestered in the interior while the hydrophilic residues are exposed to the solvent. The protein can be unfolded (denatured) by heating, changing the pH, or adding reagents such as detergents, urea, or guanidine hydrochloride. Myoglobin is a common model protein used to investigate the structural changes that can occur with pH titration. Myoglobin is an extremely compact heme protein (MW ~ 17,800), found primarily in cardiac and red skeletal muscles, which functions in the storage of oxygen and facilitates the transport of oxygen to the mitochondria for oxidative phosphorylation.
Myoglobin is particularly abundant in diving mammals including the whale, seal, and porpoise that are able to remain submerged for long periods due to the storage of oxygen by muscle myoglobin. Myoglobin consists of a single polypeptide chain of about 153 amino acids. Approximately 70% of the main chain is folded into eight major, right-handed alpha helices. The majority of the rest of the chain forms turns between helices devoid of symmetry. Four of the helices are terminated with a proline residue, whose five-membered ring does not fit within a straight stretch of the alpha helix, thereby disrupting it. Hydrophobic attractions are the driving force behind protein folding.
The conformations of the three physiologically pertinent forms of myoglobin-deoxymyoglobin, oxymyoglobin, and metmyoglobin (ferrimyoglobin)-are very similar except at the sixth coordination position. Methods including fluorescence, circular dichroism (CD), nuclear magnetic resonance (NMR), and electrometric titrations have been utilized to investigate the conformations of myoglobin. After loss of the heme, unfolding occurs in two stages: partial unfolding of the native apoprotein to a molten globule intermediate and then complete disruption of all the helical segments.
In the structural model for myoglobin denaturation, ferrimyoglobin oxidizes and loses heme, yielding apomyoglobin in the N state. The heme group dissociates from the protein at very low pH. The N-form observed under neutral and mildly acidic conditions (pH 4.5 – 7.0) has ~80% alpha helical content. Likewise, the N-form observed under neutral and mildly acidic conditions has ~55% alpha helix content. The B, C, and E helices which make up the heme pocket then unfold to give the molten globule state in which the A, G, and H helices are still intact. The last step is the conversion of the intermediate to the completely unfolded U state. The U-forms have a small residual alpha helix content and high intrinsic viscosity under highly acidic conditions (pH <2.0) indicative of a random-coil conformation. The resistance of holomyoglobin to denaturation is a function of both the intrinsic stability of the apoprotein tertiary structure and the strength of the interactions with the prosthetic group.
Experimental

An 18μg/mL solution of myoglobin was prepared by dissolving horse skeletal muscle myoglobin in deionized water. Chemical denaturation of the protein was initiated by the addition of 0.1M Sulfuric Acid using an automated titrator (ATS-429). The protein unfolding was followed using a JASCO J-1500 CD Spectrometer. The sample was contained in a 1cm quartz cuvette using a magnetic stirrer. Myoglobin CD spectra were automatically measured at 0.05 mL intervals using Spectra Manager Software. (Figure 1) The totally automated study was completed in just under an hour. CD spectra were collected from 260/180 nm with a data pitch of 0.1 nm. A band width of 1 nm was used with a detector response time of 4 sec. and scanning speed of 50nm/min. Data was analyzed using Jasco Interval Scan Analysis.
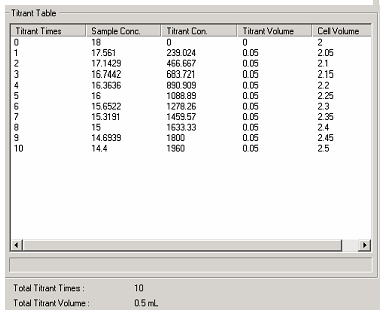
the change in sample and titrant concentration that will
occur with each subsequent titration.
Keywords
myoglobin secondary structure
Results
The pH Titration
Sulfuric acid was chosen for the titration since the chloride ions in an acid like HCl interfere with the CD spectrum. Several concentrations of acid were evaluated with 0.1M chosen. Concentrations higher than this caused immediate denaturation upon the first delivery. As shown in Figure 1, the Spectra Manager software automatically calculates the concentration of both the titrant (H2SO4) and the myoglobin after each aliquot. The software can perform the calculation to maintain a constant volume in the cuvette.
As shown in Figure 2, the CD of the myoglobin changed as the pH of the solution decreased. The intensity of the band centered at 193nm began a gradual decrease until 0.5mL of acid had been added and the pH lowered to approximately 2. At this point the helices which comprise the heme pocket have unfolded and lost some of their alpha helical shape.
Figure 3 shows a screen shot of the cross-sectional analysis program. Using this feature a wavelength can be selected and the change in the CD signal followed with increasing concentration of the titrant. Using this feature one can clearly see that the protein began to dissociate at an acid concentration of 125ppm and that the denaturation is finished once the acid concentration reaches 200ppm. This clearly demonstrates the loss of the alpha helical structure of the myoglobin.
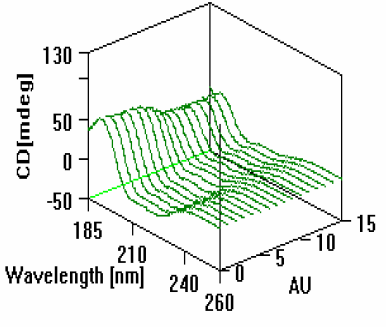
changes that occur with pH titration.
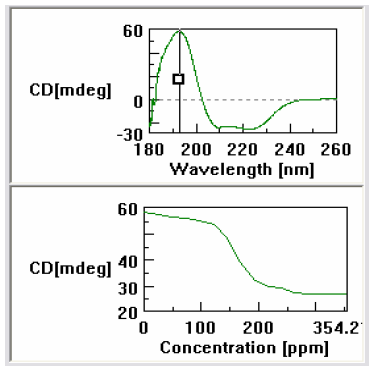
changes that occur at 193nm.
Featured Products:

Myoglobin Secondary Structure Measurement using Circular Dichroism with Automated pH Titration
Introduction
Effects of pH on Myoglobin Secondary Structure
In this application note the effects of pH on myoglobin secondary structure is measured using circular dichroism with automated titration. Circular dichroism spectroscopy (CD) is very sensitive to the secondary structure of poly-peptides and proteins. Circular dichroism is a form of light absorption spectroscopy that measures the difference in absorbance of right and left-circularly polarized light (rather than the commonly used absorbance of isotropic light) by a substance. It has been shown that CD spectra between 260 and approximately 180 nm can be analyzed for the different secondary structure types: alpha helix, beta sheet, beta turn, random coils, etc. Secondary structure determination by CD is reported to achieve accuracies of 0.97 for helices, 0.75 for beta sheet, 0.50 for turns, and 0.89 for other structural motifs.
A protein can be considered as a hetero-polymer, often hundreds of units long, composed of twenty basic amino acids. The sequence of amino acids dictates the well-defined native 3D structure, which often includes helix, sheet, turn, and random coil regions. Although the backbone of amino acids is covalently linked, usually non-covalent interactions are responsible for the folded 3D structure. In the native protein, the hydrophobic amino acid residues are generally sequestered in the interior while the hydrophilic residues are exposed to the solvent. The protein can be unfolded (denatured) by heating, changing the pH, or adding reagents such as detergents, urea, or guanidine hydrochloride. Myoglobin is a common model protein used to investigate the structural changes that can occur with pH titration. Myoglobin is an extremely compact heme protein (MW ~ 17,800), found primarily in cardiac and red skeletal muscles, which functions in the storage of oxygen and facilitates the transport of oxygen to the mitochondria for oxidative phosphorylation.
Myoglobin is particularly abundant in diving mammals including the whale, seal, and porpoise that are able to remain submerged for long periods due to the storage of oxygen by muscle myoglobin. Myoglobin consists of a single polypeptide chain of about 153 amino acids. Approximately 70% of the main chain is folded into eight major, right-handed alpha helices. The majority of the rest of the chain forms turns between helices devoid of symmetry. Four of the helices are terminated with a proline residue, whose five-membered ring does not fit within a straight stretch of the alpha helix, thereby disrupting it. Hydrophobic attractions are the driving force behind protein folding.
The conformations of the three physiologically pertinent forms of myoglobin-deoxymyoglobin, oxymyoglobin, and metmyoglobin (ferrimyoglobin)-are very similar except at the sixth coordination position. Methods including fluorescence, circular dichroism (CD), nuclear magnetic resonance (NMR), and electrometric titrations have been utilized to investigate the conformations of myoglobin. After loss of the heme, unfolding occurs in two stages: partial unfolding of the native apoprotein to a molten globule intermediate and then complete disruption of all the helical segments.
In the structural model for myoglobin denaturation, ferrimyoglobin oxidizes and loses heme, yielding apomyoglobin in the N state. The heme group dissociates from the protein at very low pH. The N-form observed under neutral and mildly acidic conditions (pH 4.5 – 7.0) has ~80% alpha helical content. Likewise, the N-form observed under neutral and mildly acidic conditions has ~55% alpha helix content. The B, C, and E helices which make up the heme pocket then unfold to give the molten globule state in which the A, G, and H helices are still intact. The last step is the conversion of the intermediate to the completely unfolded U state. The U-forms have a small residual alpha helix content and high intrinsic viscosity under highly acidic conditions (pH <2.0) indicative of a random-coil conformation. The resistance of holomyoglobin to denaturation is a function of both the intrinsic stability of the apoprotein tertiary structure and the strength of the interactions with the prosthetic group.
Experimental

An 18μg/mL solution of myoglobin was prepared by dissolving horse skeletal muscle myoglobin in deionized water. Chemical denaturation of the protein was initiated by the addition of 0.1M Sulfuric Acid using an automated titrator (ATS-429). The protein unfolding was followed using a JASCO J-1500 CD Spectrometer. The sample was contained in a 1cm quartz cuvette using a magnetic stirrer. Myoglobin CD spectra were automatically measured at 0.05 mL intervals using Spectra Manager Software. (Figure 1) The totally automated study was completed in just under an hour. CD spectra were collected from 260/180 nm with a data pitch of 0.1 nm. A band width of 1 nm was used with a detector response time of 4 sec. and scanning speed of 50nm/min. Data was analyzed using Jasco Interval Scan Analysis.

the change in sample and titrant concentration that will
occur with each subsequent titration.
Results
The pH Titration
Sulfuric acid was chosen for the titration since the chloride ions in an acid like HCl interfere with the CD spectrum. Several concentrations of acid were evaluated with 0.1M chosen. Concentrations higher than this caused immediate denaturation upon the first delivery. As shown in Figure 1, the Spectra Manager software automatically calculates the concentration of both the titrant (H2SO4) and the myoglobin after each aliquot. The software can perform the calculation to maintain a constant volume in the cuvette.
As shown in Figure 2, the CD of the myoglobin changed as the pH of the solution decreased. The intensity of the band centered at 193nm began a gradual decrease until 0.5mL of acid had been added and the pH lowered to approximately 2. At this point the helices which comprise the heme pocket have unfolded and lost some of their alpha helical shape.
Figure 3 shows a screen shot of the cross-sectional analysis program. Using this feature a wavelength can be selected and the change in the CD signal followed with increasing concentration of the titrant. Using this feature one can clearly see that the protein began to dissociate at an acid concentration of 125ppm and that the denaturation is finished once the acid concentration reaches 200ppm. This clearly demonstrates the loss of the alpha helical structure of the myoglobin.

changes that occur with pH titration.

changes that occur at 193nm.
Keywords
myoglobin secondary structure

 Download This Application
Download This Application

