Analysis of Primary and Secondary Amino Acids by Online Pre-Column Derivatization with Ortho-Ohthalaldehyde (OPA) and 9-Fluorenylmethyl Chloroformate (FMOC) Using a Rapid High-Performance Liquid Chromatography (RHPLC) System
October 21, 2024
Introduction
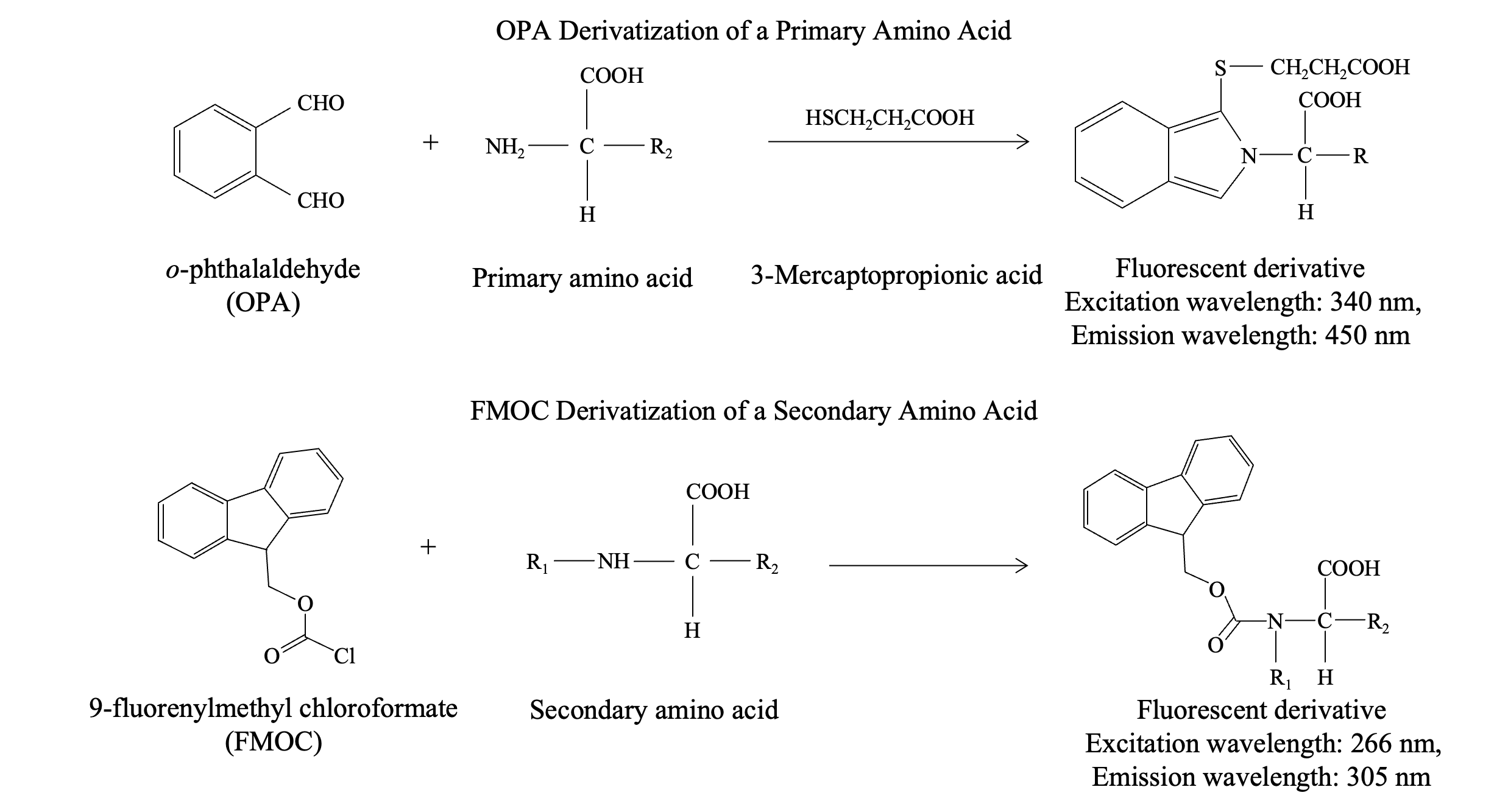
The pre-column derivatization method for amino acid analysis involves chemically modifying amino acids before separation on a column, such as a C18 column, followed by fluorescence detection. A commonly used reagent for derivatizing primary amino acids is o-phthalaldehyde (OPA); however, OPA does not react with secondary amino acids, such as proline, which plays a crucial role in proteins and is prevalent in various foods and beverages. Consequently, there are instances where analyzing secondary amino acids is essential. For this purpose, 9-fluorenylmethyl chloroformate (FMOC) is a commonly used reagent.
To overcome these limitations, a dual-reagent pre-column derivatization method can be employed using both OPA and FMOC. In this method, OPA is used to derivatize primary amino acids, while FMOC is used to derivatize secondary amino acids, such as proline. Subsequently, each amino acid can then be detected as a fluorescent derivative using a fluorescence detector.
Although the fluorescent derivatives of OPA and FMOC exhibit different excitation and emission wavelengths, simultaneous analysis can be achieved by automatically switching the detection wavelength. Furthermore, the control parameters of the autosampler can facilitate the automation of the entire derivatization process, including dispensing, mixing, and agitation of the reaction solutions.
In this application note, a standard sample containing 22 amino acids was successfully separated and analyzed using the online pre-column derivatization method utilizing OPA and FMOC under RHPLC conditions.
Experimental
RHPLC System Configuration
| Pump | PU-4185-Binary |
| Autosampler | AS-4150* |
| Column Oven | CO-4060 |
| Fluorescence Detector | FP-4020* |
| *equipped with optional units | |
RHPLC Conditions
| Column | Unifinepak C18 (3.0 mm I.D. x 75 mm L, 1.9 µm) |
| Mobile Phase A | 20 mM phosphate potassium buffer (adjusted to pH 6.9) |
| Mobile Phase B (%) | Acetonitrile/Methanol/Water (45/40/15) |
| Gradient Profile | Mobile Phase A / Mobile Phase B = 89/11 (0.0 min) → 87/13 (3.0 min) → 69/31 (5.0 min) → 63/37 (7.0 min) → 30/70 (11.0 min) → 0/100 (11.5 min) → 0/100 (14.5 min) →89/11 (15.0 min), 1 cycle; 20 minutes |
| Flow Rate | 0.8 mL/minute |
| Reagents | 3-mercaptopropionic acid (3-MPA) solution (10 µL 3-MPA in 10 mL of 0.1 mol/L borate buffer, pH 9.2) OPA solution (20 mg OPA in 10 mL of 0.1 mol/L borate buffer, pH 9.2) FMOC solution (2 mg FMOC in 10 mL of acetonitrile) |
| Column Temperature | 35 ºC |
| Wavelengths | Ex. 340 nm, Em. 450 nm → Ex. 266 nm, Em. 305 nm (10.5 min) (Gain x10) |
| Injection Volume | 1 µL |
| Standard Solution | 22 amino acids in 0.1 mol/L hydrochloric acid (HCl) |
OPA and FMOC Derivatization Reactions
Keywords
Rapid High-Performance Liquid Chromatography, RHPLC, Fluorescence Detection, Pre-column Derivatization, ortho-phthalaldehyde, o-phthalaldehyde, OPA, 9-fluorenylmethyl chloroformate, FMOC, 3-mercaptopropionic acid, 3-MPA, Amino Acids, Primary Amino Acids, Secondary Amino Acids, Unifinepak C18
Results
RHPLC System’s Performance Verification
To verify the RHPLC system’s performance, a standard solution that contains 22 different amino acids was analyzed.
Figure 1 shows the chromatogram of an amino acid standard solution that contains 100 µmol/L of 22 different amino acids.
1: Aspartic acid, 2: Glutamic acid, 3: Asparagine, 4: Serine, 5: Glutamine, 6: Histidine, 7: Glycine, 8: Threonine, 9: Citrulline, 10: Arginine, 11: Alanine, 12: GABA, 13: Tyrosine, 14: Cystine, 15: Valine, 16: Methionine, 17: Tryptophan, 18: Phenylalanine, 19: Isoleucine, 20: Leucine, 21: Lysine, 22: Proline
Table 1 shows the reproducibility of the retention times and the peak areas for each of the 22 amino acids in the standard solution analyzed using the pre-column derivatization method. The results confirm excellent reproducibility for all 22 amino acid peaks, with retention times being less than 0.2% RSD and peak areas being less than 3% RSD.
Table 1. Reproducibility and Detection Limit for the 22 Amino Acids in the Standard Solution (100 µmol/L, n = 6)
| Amino Acid Peak | Detection Limit [fmol]* (S/N = 3) | Amino Acid Peak | Detection Limit [fmol]* (S/N = 3) |
||||
| Retention Time | Peak Area | Retention Time | Peak Area | ||||
| (1) Aspartic Acid | 0.00 | 0.23 | 130 | (12) GABA | 0.02 | 0.59 | 124 |
| (2) Glutamic Acid | 0.15 | 0.12 | 158 | (13) Tyrosine | 0.02 | 0.27 | 110 |
| (3) Asparagine | 0.17 | 0.38 | 136 | (14) Cystine | 0.07 | 0.78 | 775 |
| (4) Serine | 0.15 | 0.32 | 153 | (15) Valine | 0.06 | 0.51 | 108 |
| (5) Glutamine | 0.16 | 0.64 | 245 | (16) Methionine | 0.15 | 0.80 | 243 |
| (6) Histidine | 0.15 | 0.79 | 298 | (17) Tryptophan | 0.15 | 0.39 | 118 |
| (7) Glycine | 0.13 | 1.04 | 200 | (18) Phenylalanine | 0.15 | 0.30 | 108 |
| (8) Threonine | 0.12 | 0.34 | 189 | (19) Isoleucine | 0.15 | 0.25 | 97 |
| (9) Citrulline | 0.08 | 0.30 | 126 | (20) Leucine | 0.15 | 0.30 | 102 |
| (10) Arginine | 0.06 | 0.31 | 116 | (21) Lysine | 0.17 | 0.62 | 297 |
| (11) Alanine | 0.03 | 0.28 | 122 | (22) Proline | 0.12 | 2.06 | 78 |
*Calculated from measuring the amino acid standard solution at 100 µmol/L.
Amino Acid Standard Solution Calibration Curve
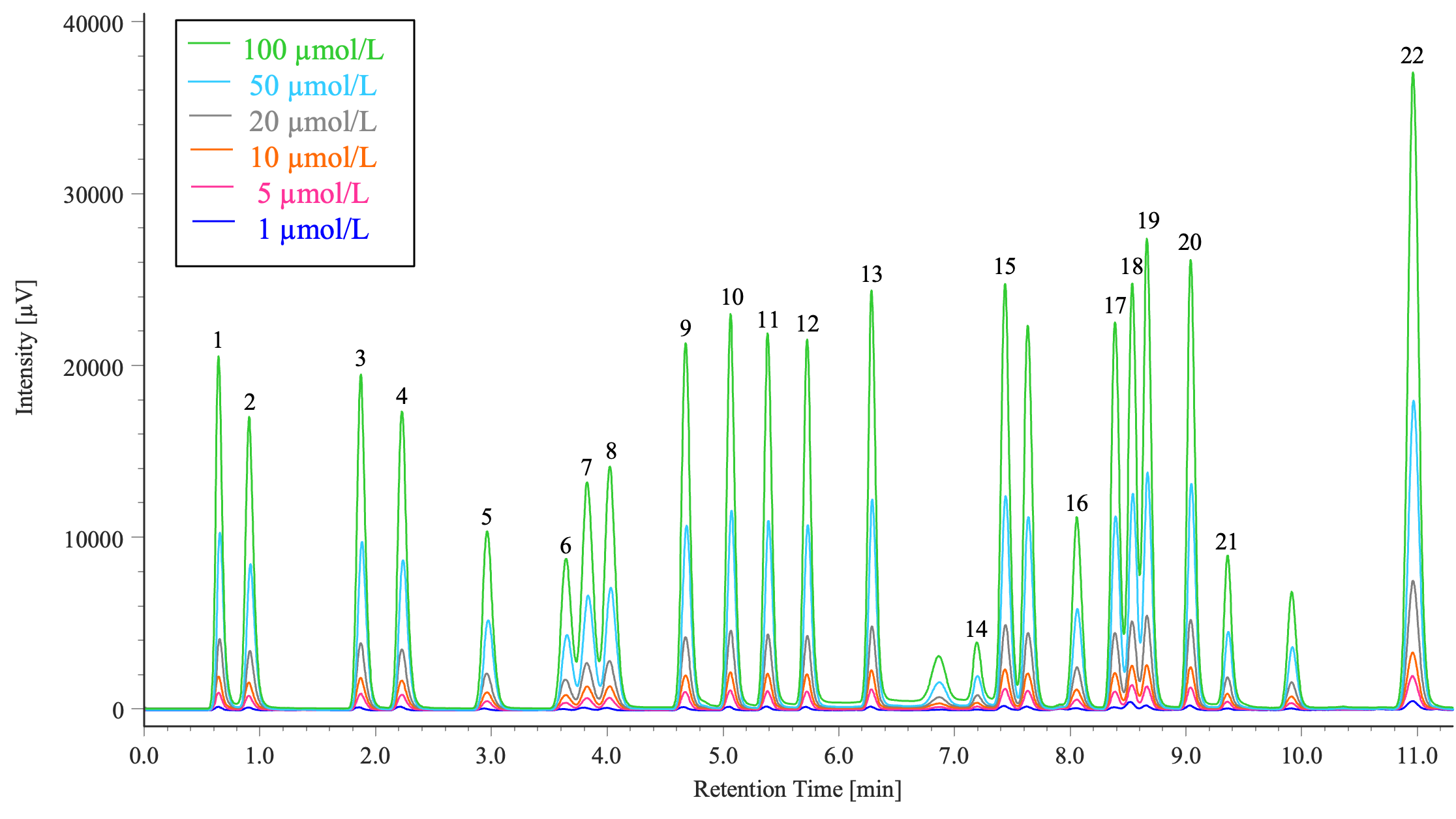
Figure 2 shows the overlaid chromatograms of six different concentrations of the amino acid standard solution that contains 22 different amino acids in the concentration range of 1 to 100 µmol/L (1, 5, 10, 20, 50, and 100 µmol/L).

1: Aspartic acid, 2: Glutamic acid, 3: Asparagine, 4: Serine, 5: Glutamine, 6: Histidine, 7: Glycine, 8: Threonine, 9: Citrulline, 10: Arginine, 11: Alanine, 12: GABA, 13: Tyrosine, 14: Cystine, 15: Valine, 16: Methionine, 17: Tryptophan, 18: Phenylalanine, 19: Isoleucine, 20: Leucine, 21: Lysine, 22: Proline
Table 2 shows the correlation coefficients for each of the 22 amino acid calibration curves analyzed using the pre-column derivatization method were all greater than or equal to r = 0.9998, indicating excellent linearity.
Table 2. Calibration Curve Correlation Coefficients for the 22 Amino Acids in the Standard Solution
| Amino Acid Peak | Correlation Coefficient | Amino Acid Peak | Correlation Coefficient |
| (1) Aspartic Acid | 1.0000 | (12) GABA | 1.0000 |
| (2) Glutamic acid | 1.0000 | (13) Tyrosine | 1.0000 |
| (3) Asparagine | 0.9999 | (14) Cystine | 0.9999 |
| (4) Serine | 1.0000 | (15) Valine | 1.0000 |
| (5) Glutamine | 0.9999 | (16) Methionine | 0.9999 |
| (6) Histidine | 1.0000 | (17) Tryptophan | 1.0000 |
| (7) Glycine | 1.0000 | (18) Phenylalanine | 0.9999 |
| (8) Threonine | 1.0000 | (19) Isoleucine | 1.0000 |
| (9) Citrulline | 0.9999 | (20) Leucine | 1.0000 |
| (10) Arginine | 0.9999 | (21) Lysine | 1.0000 |
| (11) Alanine | 1.0000 | (22) Proline | 0.9998 |
Measurement Results of Two Samples (Soy Sauce and a Sports Drink)
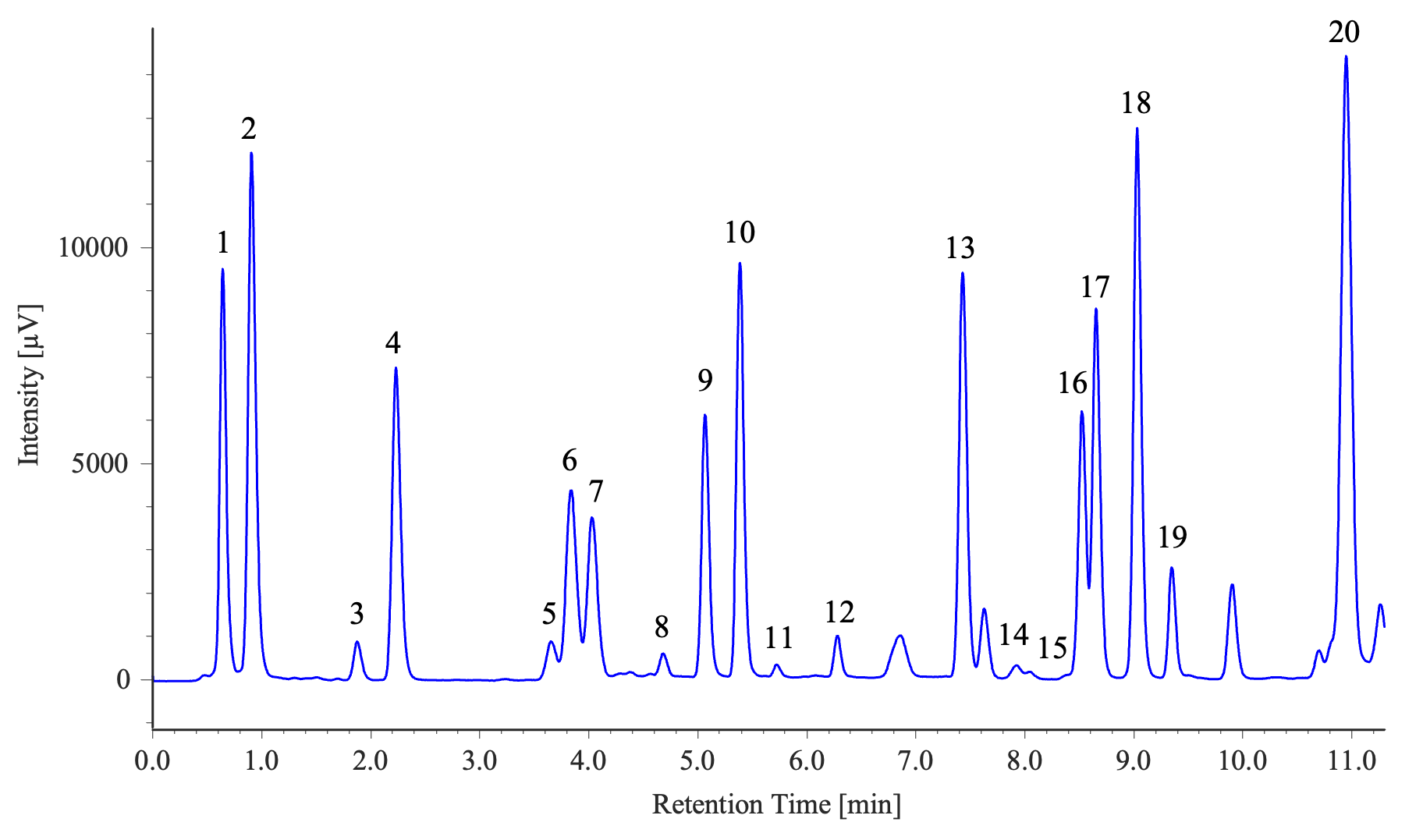
1: Aspartic acid, 2: Glutamic acid, 3: Asparagine, 4: Serine, 5: Histidine, 6: Glycine, 7: Threonine, 8: Citrulline, 9: Arginine, 10: Alanine, 11: GABA, 12: Tyrosine, 13: Valine, 14: Methionine, 15: Tryptophan, 16: Phenylalanine, 17: Isoleucine, 18: Leucine, 19: Lysine, 20: Proline
Sample Preparation Method (Soy Sauce)
1) Sample dilution by a factor of 1,000 with 0.1 M HCl.
2) Filtration through a 0.45 µm membrane filter.
3) Filtration through a 0.2 µm membrane filter.
4) Injection into the RHPLC system.

1: Aspartic acid, 2: Glutamic acid, 3: Asparagine, 4: Serine, 5: Histidine, 6: Glycine, 7: Threonine, 8: Arginine, 9: Alanine, 10: GABA, 11: Tyrosine, 12: Valine, 13: Methionine, 14: Phenylalanine, 15: Isoleucine, 16: Leucine, 17: Lysine, 18: Proline
Sample Preparation Method (Sports Drink)
1) Sample dilution by a factor of 2 with 0.1 M HCl.
2) Filtration through a 0.45 µm membrane filter.
3) Filtration through a 0.2 µm membrane filter.
4) Injection into the RHPLC system.
Automatic Pre-column Derivatization Using the AS-4150 Autosampler

Figure 5 shows the procedure for automatic pre-column derivatization using the control parameters of the AS-4150 autosampler. By utilizing this procedure, it becomes possible to perform pre-column derivatization automatically with just the preparation of samples, reagent solutions, and mixing vials.
Conclusion
This application note confirms that the measurement of samples containing primary and secondary amino acids, including proline, can be successfully separated and detected as fluorescent derivatives using the pre-column derivatization method with OPA and FMOC under RHPLC conditions.
Related Products

Analysis of Primary and Secondary Amino Acids by Online Pre-Column Derivatization with Ortho-Ohthalaldehyde (OPA) and 9-Fluorenylmethyl Chloroformate (FMOC) Using a Rapid High-Performance Liquid Chromatography (RHPLC) System
Introduction
The pre-column derivatization method for amino acid analysis involves chemically modifying amino acids before separation on a column, such as a C18 column, followed by fluorescence detection. A commonly used reagent for derivatizing primary amino acids is o-phthalaldehyde (OPA); however, OPA does not react with secondary amino acids, such as proline, which plays a crucial role in proteins and is prevalent in various foods and beverages. Consequently, there are instances where analyzing secondary amino acids is essential. For this purpose, 9-fluorenylmethyl chloroformate (FMOC) is a commonly used reagent.
To overcome these limitations, a dual-reagent pre-column derivatization method can be employed using both OPA and FMOC. In this method, OPA is used to derivatize primary amino acids, while FMOC is used to derivatize secondary amino acids, such as proline. Subsequently, each amino acid can then be detected as a fluorescent derivative using a fluorescence detector.
Although the fluorescent derivatives of OPA and FMOC exhibit different excitation and emission wavelengths, simultaneous analysis can be achieved by automatically switching the detection wavelength. Furthermore, the control parameters of the autosampler can facilitate the automation of the entire derivatization process, including dispensing, mixing, and agitation of the reaction solutions.
In this application note, a standard sample containing 22 amino acids was successfully separated and analyzed using the online pre-column derivatization method utilizing OPA and FMOC under RHPLC conditions.
Experimental
RHPLC System Configuration
| Pump | PU-4185-Binary |
| Autosampler | AS-4150* |
| Column Oven | CO-4060 |
| Fluorescence Detector | FP-4020* |
| *equipped with optional units | |
RHPLC Conditions
| Column | Unifinepak C18 (3.0 mm I.D. x 75 mm L, 1.9 µm) |
| Mobile Phase A | 20 mM phosphate potassium buffer (adjusted to pH 6.9) |
| Mobile Phase B (%) | Acetonitrile/Methanol/Water (45/40/15) |
| Gradient Profile | Mobile Phase A / Mobile Phase B = 89/11 (0.0 min) → 87/13 (3.0 min) → 69/31 (5.0 min) → 63/37 (7.0 min) → 30/70 (11.0 min) → 0/100 (11.5 min) → 0/100 (14.5 min) →89/11 (15.0 min), 1 cycle; 20 minutes |
| Flow Rate | 0.8 mL/minute |
| Reagents | 3-mercaptopropionic acid (3-MPA) solution (10 µL 3-MPA in 10 mL of 0.1 mol/L borate buffer, pH 9.2) OPA solution (20 mg OPA in 10 mL of 0.1 mol/L borate buffer, pH 9.2) FMOC solution (2 mg FMOC in 10 mL of acetonitrile) |
| Column Temperature | 35 ºC |
| Wavelengths | Ex. 340 nm, Em. 450 nm → Ex. 266 nm, Em. 305 nm (10.5 min) (Gain x10) |
| Injection Volume | 1 µL |
| Standard Solution | 22 amino acids in 0.1 mol/L hydrochloric acid (HCl) |
OPA and FMOC Derivatization Reactions
Results
RHPLC System’s Performance Verification
To verify the RHPLC system’s performance, a standard solution that contains 22 different amino acids was analyzed.
Figure 1 shows the chromatogram of an amino acid standard solution that contains 100 µmol/L of 22 different amino acids.

1: Aspartic acid, 2: Glutamic acid, 3: Asparagine, 4: Serine, 5: Glutamine, 6: Histidine, 7: Glycine, 8: Threonine, 9: Citrulline, 10: Arginine, 11: Alanine, 12: GABA, 13: Tyrosine, 14: Cystine, 15: Valine, 16: Methionine, 17: Tryptophan, 18: Phenylalanine, 19: Isoleucine, 20: Leucine, 21: Lysine, 22: Proline
Table 1 shows the reproducibility of the retention times and the peak areas for each of the 22 amino acids in the standard solution analyzed using the pre-column derivatization method. The results confirm excellent reproducibility for all 22 amino acid peaks, with retention times being less than 0.2% RSD and peak areas being less than 3% RSD.
Table 1. Reproducibility and Detection Limit for the 22 Amino Acids in the Standard Solution (100 µmol/L, n = 6)
| Amino Acid Peak | Detection Limit [fmol]* (S/N = 3) | Amino Acid Peak | Detection Limit [fmol]* (S/N = 3) |
||||
| Retention Time | Peak Area | Retention Time | Peak Area | ||||
| (1) Aspartic Acid | 0.00 | 0.23 | 130 | (12) GABA | 0.02 | 0.59 | 124 |
| (2) Glutamic Acid | 0.15 | 0.12 | 158 | (13) Tyrosine | 0.02 | 0.27 | 110 |
| (3) Asparagine | 0.17 | 0.38 | 136 | (14) Cystine | 0.07 | 0.78 | 775 |
| (4) Serine | 0.15 | 0.32 | 153 | (15) Valine | 0.06 | 0.51 | 108 |
| (5) Glutamine | 0.16 | 0.64 | 245 | (16) Methionine | 0.15 | 0.80 | 243 |
| (6) Histidine | 0.15 | 0.79 | 298 | (17) Tryptophan | 0.15 | 0.39 | 118 |
| (7) Glycine | 0.13 | 1.04 | 200 | (18) Phenylalanine | 0.15 | 0.30 | 108 |
| (8) Threonine | 0.12 | 0.34 | 189 | (19) Isoleucine | 0.15 | 0.25 | 97 |
| (9) Citrulline | 0.08 | 0.30 | 126 | (20) Leucine | 0.15 | 0.30 | 102 |
| (10) Arginine | 0.06 | 0.31 | 116 | (21) Lysine | 0.17 | 0.62 | 297 |
| (11) Alanine | 0.03 | 0.28 | 122 | (22) Proline | 0.12 | 2.06 | 78 |
*Calculated from measuring the amino acid standard solution at 100 µmol/L.
Amino Acid Standard Solution Calibration Curve
Figure 2 shows the overlaid chromatograms of six different concentrations of the amino acid standard solution that contains 22 different amino acids in the concentration range of 1 to 100 µmol/L (1, 5, 10, 20, 50, and 100 µmol/L).

1: Aspartic acid, 2: Glutamic acid, 3: Asparagine, 4: Serine, 5: Glutamine, 6: Histidine, 7: Glycine, 8: Threonine, 9: Citrulline, 10: Arginine, 11: Alanine, 12: GABA, 13: Tyrosine, 14: Cystine, 15: Valine, 16: Methionine, 17: Tryptophan, 18: Phenylalanine, 19: Isoleucine, 20: Leucine, 21: Lysine, 22: Proline
Table 2 shows the correlation coefficients for each of the 22 amino acid calibration curves analyzed using the pre-column derivatization method were all greater than or equal to r = 0.9998, indicating excellent linearity.
Table 2. Calibration Curve Correlation Coefficients for the 22 Amino Acids in the Standard Solution
| Amino Acid Peak | Correlation Coefficient | Amino Acid Peak | Correlation Coefficient |
| (1) Aspartic Acid | 1.0000 | (12) GABA | 1.0000 |
| (2) Glutamic acid | 1.0000 | (13) Tyrosine | 1.0000 |
| (3) Asparagine | 0.9999 | (14) Cystine | 0.9999 |
| (4) Serine | 1.0000 | (15) Valine | 1.0000 |
| (5) Glutamine | 0.9999 | (16) Methionine | 0.9999 |
| (6) Histidine | 1.0000 | (17) Tryptophan | 1.0000 |
| (7) Glycine | 1.0000 | (18) Phenylalanine | 0.9999 |
| (8) Threonine | 1.0000 | (19) Isoleucine | 1.0000 |
| (9) Citrulline | 0.9999 | (20) Leucine | 1.0000 |
| (10) Arginine | 0.9999 | (21) Lysine | 1.0000 |
| (11) Alanine | 1.0000 | (22) Proline | 0.9998 |
Measurement Results of Two Samples (Soy Sauce and a Sports Drink)

1: Aspartic acid, 2: Glutamic acid, 3: Asparagine, 4: Serine, 5: Histidine, 6: Glycine, 7: Threonine, 8: Citrulline, 9: Arginine, 10: Alanine, 11: GABA, 12: Tyrosine, 13: Valine, 14: Methionine, 15: Tryptophan, 16: Phenylalanine, 17: Isoleucine, 18: Leucine, 19: Lysine, 20: Proline
Sample Preparation Method (Soy Sauce)
1) Sample dilution by a factor of 1,000 with 0.1 M HCl.
2) Filtration through a 0.45 µm membrane filter.
3) Filtration through a 0.2 µm membrane filter.
4) Injection into the RHPLC system.

1: Aspartic acid, 2: Glutamic acid, 3: Asparagine, 4: Serine, 5: Histidine, 6: Glycine, 7: Threonine, 8: Arginine, 9: Alanine, 10: GABA, 11: Tyrosine, 12: Valine, 13: Methionine, 14: Phenylalanine, 15: Isoleucine, 16: Leucine, 17: Lysine, 18: Proline
Sample Preparation Method (Sports Drink)
1) Sample dilution by a factor of 2 with 0.1 M HCl.
2) Filtration through a 0.45 µm membrane filter.
3) Filtration through a 0.2 µm membrane filter.
4) Injection into the RHPLC system.
Automatic Pre-column Derivatization Using the AS-4150 Autosampler

Figure 5 shows the procedure for automatic pre-column derivatization using the control parameters of the AS-4150 autosampler. By utilizing this procedure, it becomes possible to perform pre-column derivatization automatically with just the preparation of samples, reagent solutions, and mixing vials.
Conclusion
This application note confirms that the measurement of samples containing primary and secondary amino acids, including proline, can be successfully separated and detected as fluorescent derivatives using the pre-column derivatization method with OPA and FMOC under RHPLC conditions.
Keywords
Rapid High-Performance Liquid Chromatography, RHPLC, Fluorescence Detection, Pre-column Derivatization, ortho-phthalaldehyde, o-phthalaldehyde, OPA, 9-fluorenylmethyl chloroformate, FMOC, 3-mercaptopropionic acid, 3-MPA, Amino Acids, Primary Amino Acids, Secondary Amino Acids, Unifinepak C18

 Download This Application
Download This Application