Analysis of Protein Hydrolysate Amino Acids using OPA Post-column Derivatization by Quaternary Low Pressure Gradient System
January 5, 2024
Introduction
Amino acid analysis has been applied to several categories such as food, medicine, protein science and metabolome study, and is an important measurement technique.
An amino acid analysis system using a low pressure gradient unit with OPA post column derivatization provides excellent repeatability and good separation in a short analysis time.
The analysis results of food analysis and amino acid composition analysis of protein using this amino acid analysis system are reported.

Experimental
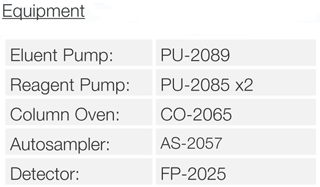
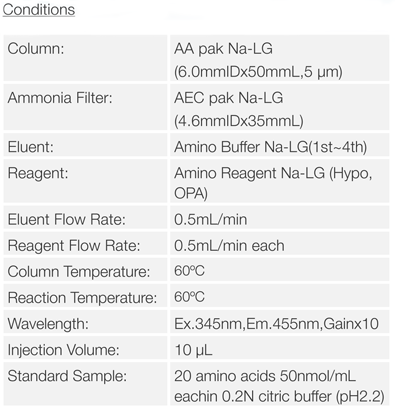
Schematic Diagram
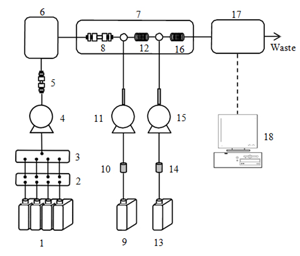
- Eluent (Amino Buffer Na-LG(1st~4th))
- Degasser
- Low pressure gradient unit
- Eluent pump
- Ammonia filter (AECpak Na-LG)
- Autosampler
- Column oven
- Column (AApak Na-LG)
- Reagent 1 (Amino Reagent Na-LG Hypo)
- Airtrap 1
- Reagent pump 1(Hypo)
- Reaction coil 1
- Reagent 2 (Amino Reagent Na-LGOPA)
- Airtrap 2
- Reagent pump 2 (OPA)
- Reaction coil 2
- Fluorescence detector
- Chromatography data system(ChromNAV)
Keywords
430027H
Results
Figure 1 shows the chromatogram of the standard mixture of 20 kinds of amino acids.The sample was well separated within 45 minutes (1 cycle 60 min).
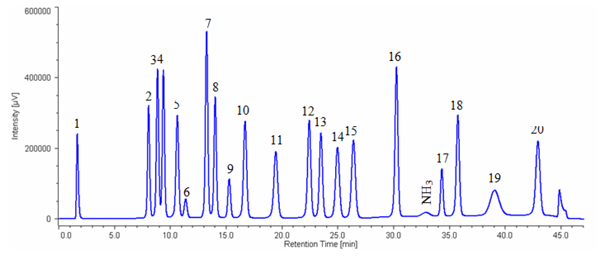
1: Cysteic acid, 2: Asparatic acid, 3: Threonine, 4: Serine, 5: Glutamicacid, 6: Proline, 7: Glycine, 8: Alanine, 9: Cystine, 10: Valine, 11: Methionine, 12: Isoleucine, 13: Leucine, 14: Tyrosine, 15: Phenylalanine, 16: GABA, 17: Lysine, 18: Histidine, 19: Tryptophan, 20: Arginine
Figure 2 shows the chromatogram of a sports drink while figure 3 shows the chromatogram of white wine containing GABA.
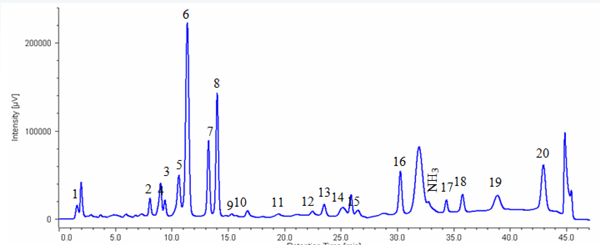
1:Cysteic acid, 2:Asparatic acid, 3:Threonine, 4:Serine, 5: Glutamicacid, 6:Proline, 7: Glycine, 8: Alanine, 9: Cystine,10: Valine,11: Methionine,12: Isoleucine,13: Leucine,14: Tyrosine,15: Phenylalanine, 16:GABA, Lysine,18: Histidine,19: Tryptophan, 20: Arginine
Sample preparation:
Sports drink was diluted 150-fold by 0.2 N citric acid buffer (pH2.2) and then filtrated using 0.45 μm membrane filter.
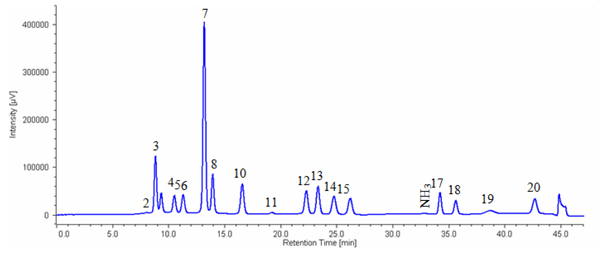
2: Asparatic acid, 3: Threonine, 4: Serine, 5: Glutamicacid, 6: Proline, 7: Glycine, 8: Alanine, 10: Valine, 11: Methionine, 12: Isoleucine, 13: Leucine, 14: Tyrosine, 15: Phenylalanine, 17: Lysine, 18:Histidine, 19: Tryptophan, 20: Arginine
Sample preparation: White wineincluding GABA was diluted 10-fold by 0.2N citric acid buffer (pH 2.2) and then filtrated using 0.45 μmmembrane filter.
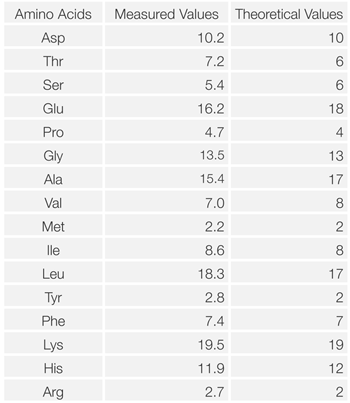
Figure 4 shows the chromatogram of hydrolyzed myoglobin (horse skeletal muscle). Table 1 shows the results of amino acid composition compared with theoretical value (referred to Japan Bio chemistry DataBook l ,Tokyo Kagaku Dojin). It was confirmed that the amino acid composi tion calculated from this measurement was in good agreement with the theoretical value.
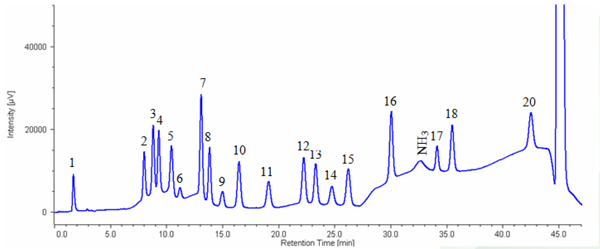
1: Cysteic acid, 2: Asparatic acid, 3: Threonine, 4: Serine, 5: Glutamicacid, 6: Proline, 7: Glycine, 8: Alanine, 9: Cystine, 10: Valine, 11: Methionine, 12: Isoleucine, 13: Leucine, 14: Tyrosine, 15: Phenylalanine, 16: GABA, 17: Lysine, 18: Histidine, 20: Arginine
Sample preparation:
1. Myoglobin was diluted to 200μg/mL by ultra pure water. 2.The solution filled a 20 μL sample tube and then dried up by centrifugal evaporator. 3. Sample tube was set in vessel for hydrolysis and 0.3 mL of hydrochloric acid was added to thevessel. 4. The sample was heated for 24 hours under vacuumed condition. 5. The sample was hydrolyzed and the residual chlorine was removed using vacuum pump. 6.500μL of 0.2N citric acid buffer (pH2.2) was added to the sample and agitated.
Table 1 Comparison of amino acid composition of myoglobin between the measured and theoretical value.
Featured Products:

Analysis of Protein Hydrolysate Amino Acids using OPA Post-column Derivatization by Quaternary Low Pressure Gradient System
Introduction
Amino acid analysis has been applied to several categories such as food, medicine, protein science and metabolome study, and is an important measurement technique.
An amino acid analysis system using a low pressure gradient unit with OPA post column derivatization provides excellent repeatability and good separation in a short analysis time.
The analysis results of food analysis and amino acid composition analysis of protein using this amino acid analysis system are reported.

Experimental


Schematic Diagram

- Eluent (Amino Buffer Na-LG(1st~4th))
- Degasser
- Low pressure gradient unit
- Eluent pump
- Ammonia filter (AECpak Na-LG)
- Autosampler
- Column oven
- Column (AApak Na-LG)
- Reagent 1 (Amino Reagent Na-LG Hypo)
- Airtrap 1
- Reagent pump 1(Hypo)
- Reaction coil 1
- Reagent 2 (Amino Reagent Na-LGOPA)
- Airtrap 2
- Reagent pump 2 (OPA)
- Reaction coil 2
- Fluorescence detector
- Chromatography data system(ChromNAV)
Keywords
430027H
Results
Figure 1 shows the chromatogram of the standard mixture of 20 kinds of amino acids.The sample was well separated within 45 minutes (1 cycle 60 min).

1: Cysteic acid, 2: Asparatic acid, 3: Threonine, 4: Serine, 5: Glutamicacid, 6: Proline, 7: Glycine, 8: Alanine, 9: Cystine, 10: Valine, 11: Methionine, 12: Isoleucine, 13: Leucine, 14: Tyrosine, 15: Phenylalanine, 16: GABA, 17: Lysine, 18: Histidine, 19: Tryptophan, 20: Arginine
Figure 2 shows the chromatogram of a sports drink while figure 3 shows the chromatogram of white wine containing GABA.

1:Cysteic acid, 2:Asparatic acid, 3:Threonine, 4:Serine, 5: Glutamicacid, 6:Proline, 7: Glycine, 8: Alanine, 9: Cystine,10: Valine,11: Methionine,12: Isoleucine,13: Leucine,14: Tyrosine,15: Phenylalanine, 16:GABA, Lysine,18: Histidine,19: Tryptophan, 20: Arginine
Sample preparation:
Sports drink was diluted 150-fold by 0.2 N citric acid buffer (pH2.2) and then filtrated using 0.45 μm membrane filter.

2: Asparatic acid, 3: Threonine, 4: Serine, 5: Glutamicacid, 6: Proline, 7: Glycine, 8: Alanine, 10: Valine, 11: Methionine, 12: Isoleucine, 13: Leucine, 14: Tyrosine, 15: Phenylalanine, 17: Lysine, 18:Histidine, 19: Tryptophan, 20: Arginine
Sample preparation: White wineincluding GABA was diluted 10-fold by 0.2N citric acid buffer (pH 2.2) and then filtrated using 0.45 μmmembrane filter.
Figure 4 shows the chromatogram of hydrolyzed myoglobin (horse skeletal muscle). Table 1 shows the results of amino acid composition compared with theoretical value (referred to Japan Bio chemistry DataBook l ,Tokyo Kagaku Dojin). It was confirmed that the amino acid composi tion calculated from this measurement was in good agreement with the theoretical value.

1: Cysteic acid, 2: Asparatic acid, 3: Threonine, 4: Serine, 5: Glutamicacid, 6: Proline, 7: Glycine, 8: Alanine, 9: Cystine, 10: Valine, 11: Methionine, 12: Isoleucine, 13: Leucine, 14: Tyrosine, 15: Phenylalanine, 16: GABA, 17: Lysine, 18: Histidine, 20: Arginine
Sample preparation:
1. Myoglobin was diluted to 200μg/mL by ultra pure water. 2.The solution filled a 20 μL sample tube and then dried up by centrifugal evaporator. 3. Sample tube was set in vessel for hydrolysis and 0.3 mL of hydrochloric acid was added to thevessel. 4. The sample was heated for 24 hours under vacuumed condition. 5. The sample was hydrolyzed and the residual chlorine was removed using vacuum pump. 6.500μL of 0.2N citric acid buffer (pH2.2) was added to the sample and agitated.
Table 1 Comparison of amino acid composition of myoglobin between the measured and theoretical value.


 Download This Application
Download This Application

