JWMVS-529 CD Multivariate SSE Analysis Program
January 5, 2024
Introduction
The function of a protein or peptide is closely related to its structure. Since CD measurements probe amino acid structure changes, CD is widely used for the structural analysis of protein and peptide molecules formulated for pharmaceutical use. CD spectral shapes reflect the abundance ratio of secondary structure motifs in proteins and poly-peptides and secondary structure analysis of CD data provides a fast and accurate indication of change in these structures.
The JWSSE-513 Protein Secondary Structure Analysis program uses the Classical Least Squares (CLS) method, including a reference spectra set of Yang1 and Reed2. The Yang reference spectra are extracted from the CD spectra of proteins and are best suited for protein secondary structure analysis. On the other hand, the Reed reference spectra are extracted from the CD spectra of peptides and are suitable for the secondary structure analysis of peptides. The two reference spectra are separate due to the aromatic side chain residue bands that are observed in the CD spectra of proteins but not peptides.
The JWMVS-529 Multivariate SSE analysis program includes a library of 26 protein CD spectra (176-260 nm) which use a calibration model based on the spectra and created by JASCO. A Partial Least Squares (PLS) method with the latest multivariate analysis method and Principle Component Regression (PCR) method are included. In the PLS and PCR methods, the spectra rely on a few potential factor for which the concentration is determined. TO minimize the residual error of the concentration, the abundance ratio of the secondary structure is calculated. This significantly improves the analysis accuracy of the β-sheet motif, which has no strong, specific CD marker band.
This application note highlights the features of the JWSEE-513 and JWMVS-529 Secondary Structure Analysis programs.

Experimental
- PLS and PCR methods are much more precise multivariate analysis methods compared with the CLS method, which has been traditionally used for protein secondary structure analysis
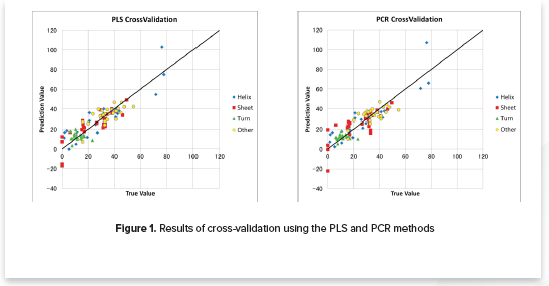
- Verifies the calibration model using cross-validation (Figure 1)
- Edits the ratio of secondary structure and reference spectra
- Validates the analysis result using a F-test
- Verification of recalculation and calculated results (GLP/GMP compliant)
- 21 CFR Part 11 compliant
- Includes CD spectra (176-260 nm) of 26 proteins with a calibration model based on those spectra
Keywords
210-CD-0018, Secondary structure analysis, multivariate, PLS method, PCR method, JWSEE-513, JWMVS-529, biochemistry
References
- Yang, J. T., Wu, C.S.C., and H. M. Martinez, Methos in Enzymology (1986), 130, 208-269.
- J. Reed and T. A. Reed, Anal. Biochem, (1997), 254, 36-40.
Featured Products:
-
J-1100
-

Highest performance with a wide range of accessories for maximum flexibility to meet complex research demands.
J-1500
-
Automated HTCD

JWMVS-529 CD Multivariate SSE Analysis Program
Introduction
The function of a protein or peptide is closely related to its structure. Since CD measurements probe amino acid structure changes, CD is widely used for the structural analysis of protein and peptide molecules formulated for pharmaceutical use. CD spectral shapes reflect the abundance ratio of secondary structure motifs in proteins and poly-peptides and secondary structure analysis of CD data provides a fast and accurate indication of change in these structures.
The JWSSE-513 Protein Secondary Structure Analysis program uses the Classical Least Squares (CLS) method, including a reference spectra set of Yang1 and Reed2. The Yang reference spectra are extracted from the CD spectra of proteins and are best suited for protein secondary structure analysis. On the other hand, the Reed reference spectra are extracted from the CD spectra of peptides and are suitable for the secondary structure analysis of peptides. The two reference spectra are separate due to the aromatic side chain residue bands that are observed in the CD spectra of proteins but not peptides.
The JWMVS-529 Multivariate SSE analysis program includes a library of 26 protein CD spectra (176-260 nm) which use a calibration model based on the spectra and created by JASCO. A Partial Least Squares (PLS) method with the latest multivariate analysis method and Principle Component Regression (PCR) method are included. In the PLS and PCR methods, the spectra rely on a few potential factor for which the concentration is determined. TO minimize the residual error of the concentration, the abundance ratio of the secondary structure is calculated. This significantly improves the analysis accuracy of the β-sheet motif, which has no strong, specific CD marker band.
This application note highlights the features of the JWSEE-513 and JWMVS-529 Secondary Structure Analysis programs.

Experimental
- PLS and PCR methods are much more precise multivariate analysis methods compared with the CLS method, which has been traditionally used for protein secondary structure analysis
- Verifies the calibration model using cross-validation (Figure 1)
- Edits the ratio of secondary structure and reference spectra
- Validates the analysis result using a F-test
- Verification of recalculation and calculated results (GLP/GMP compliant)
- 21 CFR Part 11 compliant
- Includes CD spectra (176-260 nm) of 26 proteins with a calibration model based on those spectra

Keywords
210-CD-0018, Secondary structure analysis, multivariate, PLS method, PCR method, JWSEE-513, JWMVS-529, biochemistry
References
- Yang, J. T., Wu, C.S.C., and H. M. Martinez, Methos in Enzymology (1986), 130, 208-269.
- J. Reed and T. A. Reed, Anal. Biochem, (1997), 254, 36-40.

 Download This Application
Download This Application
