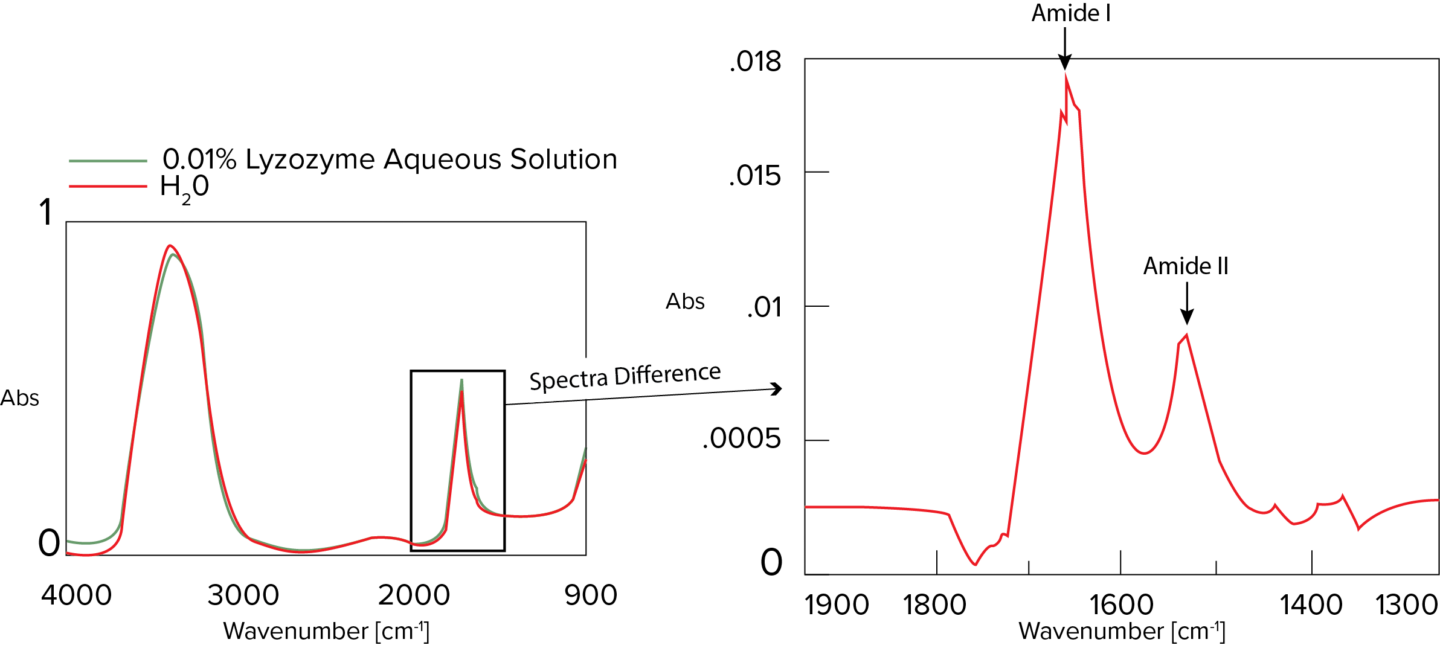
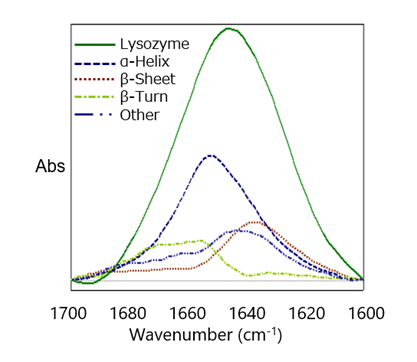
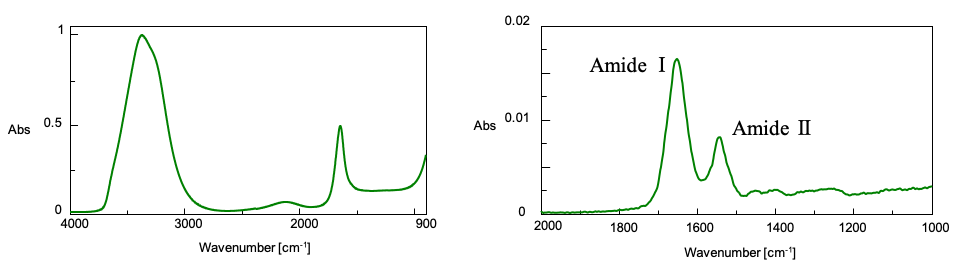
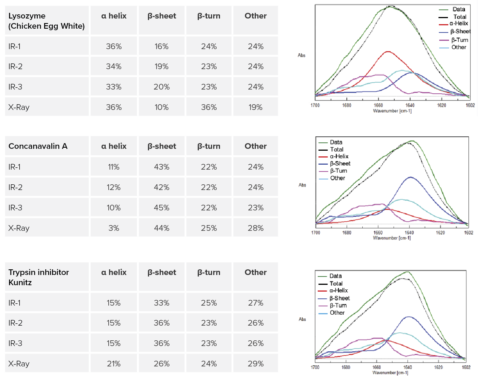
The Secondary Structure Estimation (SSE) program for FTIR includes a standard library containing 28 different types of protein spectra and that contain information about four secondary structural motifs: Helix (total number of α-helix and 3/10-helix residues), Sheet (total number of strand residues), Turn (total number of hydrogen-bonded turn residues), and Other (all other residues). The ratio of secondary structures for the proteins was calculated based on information provided by the Protein Data Bank (PDB) (as of May 2022).
Table 1. Reference Protein Molecules from the Protein Data Bank (PDB)
| Azurin | Concanavalin A | Cytochrome C | Elastase |
| Ferredoxin | Hemoglobin | Immunoglobulin G | Lysozyme |
| Myoglobin | Papain | Parvalbumin | Pepsinogen |
| Prealbumin | Ribonuclease S | Triose phosphate isomerase | α-Chymotrypsin (Bovine) |
| β-Lactoglobulin (Bovine milk) | Trypsin inhibitor (Soybean) | Elastase (Porcine pancreas) | Lysozyme (Chicken egg white) |
| α-Lactalbumin (Bovine milk) | Cytochrome C (Bovine heart) | Concanavalin A (Jack bean) | Alcohol dehydrogenase (Yeast) |
| Immunoglobulin G | Ovalbumin (Hen) | Carbonic anhydrase 1 (Human erythrocytes) | Superoxide dismutase (Cu, Zn) |